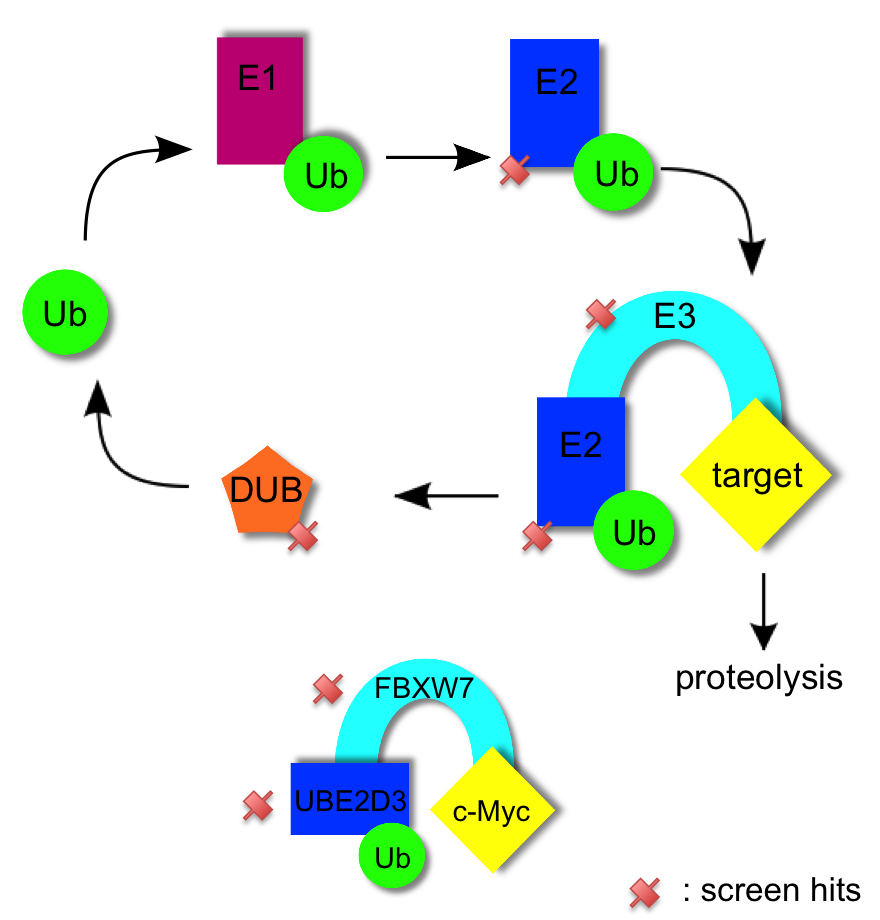
Ubiquitination is a barrier during reprogramming
Using a combination of gene ontology and gene network analysis we identified genes relevant to ubiquitination and the ubiquitin mediated proteolysis of pluripotency factors. In the canonical pathway ubiquitin is first activated (by E1 proteins), then conjugated (by E2 proteins), and then ligated (by E3 proteins) to target proteins; this ubiquitination often marks proteins for degradation. Ubuiquitin itself is liberated for reuse by de-ubiquitinating enzymes (DUBs) [1]. We identified multiple TRA-1-81+ screen hits in the ubiquitination pathway: an E2 conjugating enzyme UBE2D3 (p=0.0262), E3 ligases (FBXW7 p=0.004, NEDD4 p=0.0172, MARCH3 p=0.0152, RNF40 p=0.0044, RNF144a p=0.0002), and a DUB (USP9X p=0.0188). Thus, ubiquitination is targeted at multiple levels. Of note, ubiquitin ligase FBXW7 is significant in the TRA-1-81+ screen, as is its validated binding partner E2 enzyme UBE2D3. Fbxw7 knockdown has been shown to enhance reprogramming efficiency in mouse [2]. Similarly, in we identify the E2 conjugating enzyme UBE2D3 and its validated binding partner NEDD4 [3-4] as screen hits. Proteome array analysis shows NEDD4 to mediate ubiquitination of proteins responsible for such diverse processes as vesicle trafficking, extracellular matrix production, gene regulation, kinase activity, and protein binding events [5].
- Ciechanover, A. (2005). Proteolysis: from the lysosome to ubiquitin and the proteasome. Nature reviews. Molecular cell biology, 6(1), 79–87. doi:10.1038/nrm1552
- Buckley, S. M., Aranda-Orgilles, B., Strikoudis, A., Apostolou, E., Loizou, E., Moran-Crusio, K., Farnsworth, C. L., et al. (2012). Regulation of Pluripotency and Cellular Reprogramming by the Ubiquitin-Proteasome System. Cell Stem Cell, 11(6), 783–798. doi:10.1016/j.stem.2012.09.011
- Anan, T., Nagata, Y., Koga, H., Honda, Y., Yabuki, N., Miyamoto, C., Kuwano, a, et al. (1998). Human ubiquitin-protein ligase Nedd4: expression, subcellular localization and selective interaction with ubiquitin-conjugating enzymes. Genes to cells : devoted to molecular & cellular mechanisms, 3(11), 751–63. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9990509
- Wang, X., Shi, Y., Wang, J., Huang, G., & Jiang, X. (2008). Crucial role of the C-terminus of PTEN in antagonizing NEDD4-1-mediated PTEN ubiquitination and degradation. The Biochemical journal, 414(2), 221–9. doi:10.1042/BJ20080674
- Persaud, A., Alberts, P., Amsen, E. M., Xiong, X., Wasmuth, J., Saadon, Z., Fladd, C., et al. (2009). Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4-2 using proteome arrays. Molecular systems biology, 5(333), 333. doi:10.1038/msb.2009.85